library(glue)
library(stringi)
library(stringr)
library(ggplot2)
library(bench)I recently had to concatenate tens of millions of strings into a single column of strings. I was surprised when even my data.table code, which was something like dt[, Location := paste(chr, start, sep = "-")], was taking minutes.
So then, what is the fastest way to combine a bunch of strings?
Since I really care about this in the context of genetic data I’ll simulate strings from “Chromosomes” and “Start positions” and concatenate them into a single “Loci”. e.g. “chr1-45678”
The Candidates
Like most things in R there are a bunch of ways to complete the same task. The approaches below are a few that I could think of:
paste: base function for concatenating stringspaste0: base function for concatenating strings (paste(..., sep="")sprintf: base function for C-stylesprintfcharacter formatting of stringsstringi::stri_c:stringifunction for combining multiple character vectorsstringr::str_c:stringrfunction that wrapsstringibut conforms to tidyverse recycling and NA rulesglue::glue: String interpolation. Has to be converted to a vector after interpolation.
Create some test strings
Let’s make 1 million “chromosome-start” strings to simulate a realistic test set size.
set.seed(1234)
N <- 1e6
chroms <- sample(
paste0("chr", as.character(1:22, "X", "Y")),
size = N, replace = TRUE
)
starts <- as.character(sample.int(1e5, size = N, replace = TRUE))Time it!
The code below times the execution of each of the string joining expressions 30 times. Since glue has to be converted to a character vector after interpolation, I created a second expression without the coercion to see how much that affects the timing. It is for this reason that the check argument is set to FALSE.
# Run the benchmark
results <- bench::mark(
"paste" = paste(chroms, starts, sep = "-"),
"paste0" = paste0(chroms, "-", starts),
"sprintf" = sprintf("%s%s%s", chroms, "-", starts),
"stri_c" = stri_c(chroms, starts, sep = "-"),
"str_c" = str_c(chroms, starts, sep = "-"),
"glue" = as.character(glue("{chroms}-{starts}")),
"glue2" = glue("{chroms}-{starts}"),
check = FALSE,
memory = TRUE,
min_time = Inf,
max_iterations = 30
)
# Plot the results
autoplot(results) +
labs(
title = "Execution Time for Joining 1 Million Element Vectors",
x = "Time",
y = "Function"
) +
theme_minimal() +
theme(
plot.title = element_text(face = "bold", size = 16),
legend.position = "bottom"
)Loading required namespace: tidyr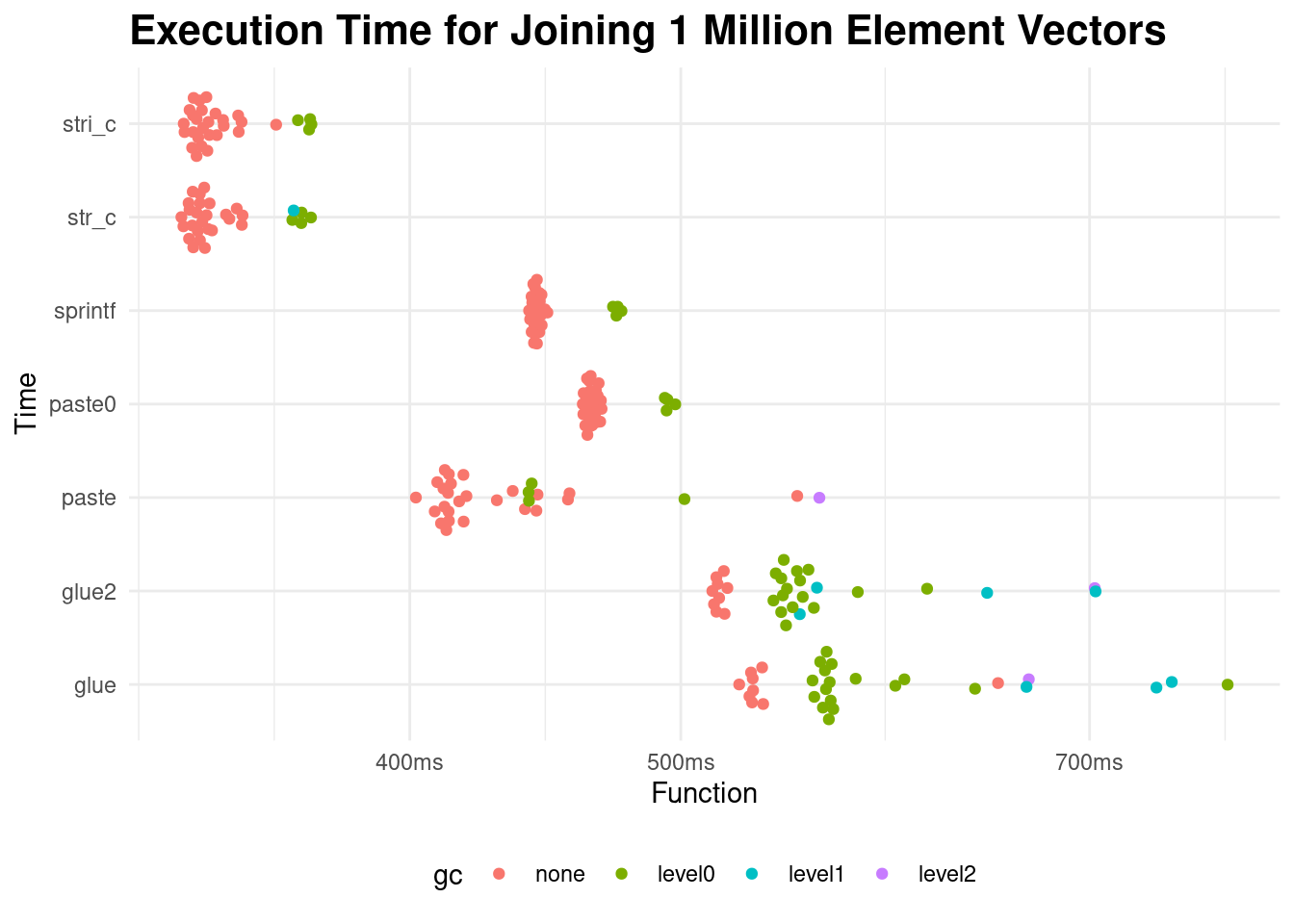
It looks like the clear winners are stringi::stri_c and stringr::str_c. This makes sense. What was surprising is that paste0 performs worse than paste with sep="-".
There’s one, somewhat hacky, solution I wanted to test. What if we were to instead write the data out as a file where the vectors are concatenated with “-” as a delimiter and then read this concatenated file back in as a single column?
data.table::fwrite & data.table::fread
Assuming more threads makes this faster(?) I’ll max out all 8 cores on my machine. Also, I doubt that this specific example will be faster on only 1 million elements so I’ll bump up the number of elements to 100 million and see how it compares against the fastest function from above.
library(data.table)
setDTthreads(percent = 100)
dt <- data.table(
Chromosome = sample(paste0("chr", as.character(1:22, "X", "Y")),
size = 1e8, replace = TRUE
),
Start = as.character(sample.int(1e5, size = 1e8, replace = TRUE))
)Write the data out using fwrite(..., sep="-") to concatenate the columns and then read them back in as a single concatenated column with fread
system.time({
fwrite(dt, file = "test.txt", sep = "-", col.names = FALSE)
fread("test.txt", col.names = "chr_start")
}) user system elapsed
57.580 1.586 34.265 And how does this compare to concatenating with stri_c
system.time(dt[, chr_start := stri_c(Chromosome, Start, sep="-")]) user system elapsed
36.670 1.280 37.952 It looks kinda crazy but maybe if you have enough space to spare and a lot of threads to throw at it then the ‘hacky’ solution might be a fast alternative for extremely large string concatenations; although the difference may not be large enough to to really matter too much in the end.